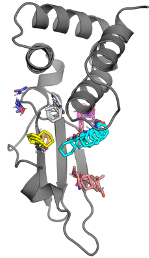Appendix C
Supplementary Material to ”Discovery of a cryptic allosteric site in Ebola’s
’undruggable’ VP35 protein”
This chapter is adapted from the following publication:
Cruz, M.A.and
Frederick, T.E.,
Singh, S., Vithani, N., Zimmerman, M.I., Porter, J.R., Moeder, K.E., Amarasinghe, G.K., and Bowman,
G.R., Discovery of a cryptic allosteric site in Ebola’s ’undruggable’ VP35 protein using simulations
and experiments. Preprint on BioRxiv https://doi.org/10.1101/2020.02.09.940510 [54]
C.1 Supplementary Material
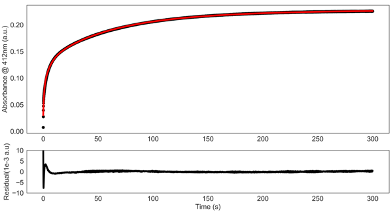
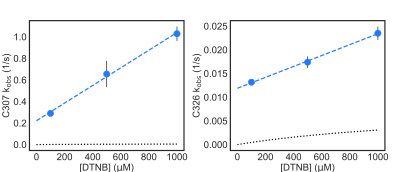
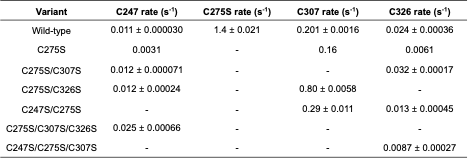
Table C.1: Characterization of the folding/unfolding of VP35’s IID used to test whether the
observed thiol labeling is due to fluctuations within the native state or global unfolding of the
protein. K is the equilibrium constant between the folded and unfolded state determined from
denaturation data, kunfold is the unfolding rate of the respective variants measured by intrinsic
tryptophan fluorescence.
| Variant | K | K(s) |
| Wild-type | 6.5710 4.010 | 0.0175 |
| C247S/C275S | 4.0110 0.810 | 0.0083 |
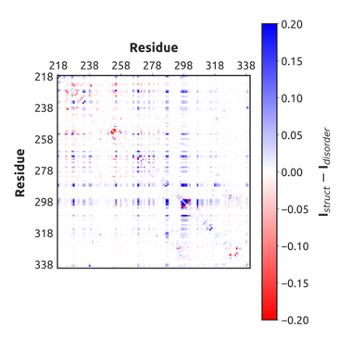
Table C.2: Intrinsic labeling rates (k)
for each cysteine residue. Intrinsic labeling rates were measured using either urea unfolded
variants containing only the specified cysteine, or peptides containing the specified cysteine
and its surrounding residues.
|
|
| Residue | kintMs |
|
|
| C247 | 0.0566 0.0007 |
|
|
| C275 | 0.00254 0.001 |
|
|
| C307 | 0.0290 0.002 |
|
|
| C326 | 0.395 0.02 |
|
|
| |