Appendix B
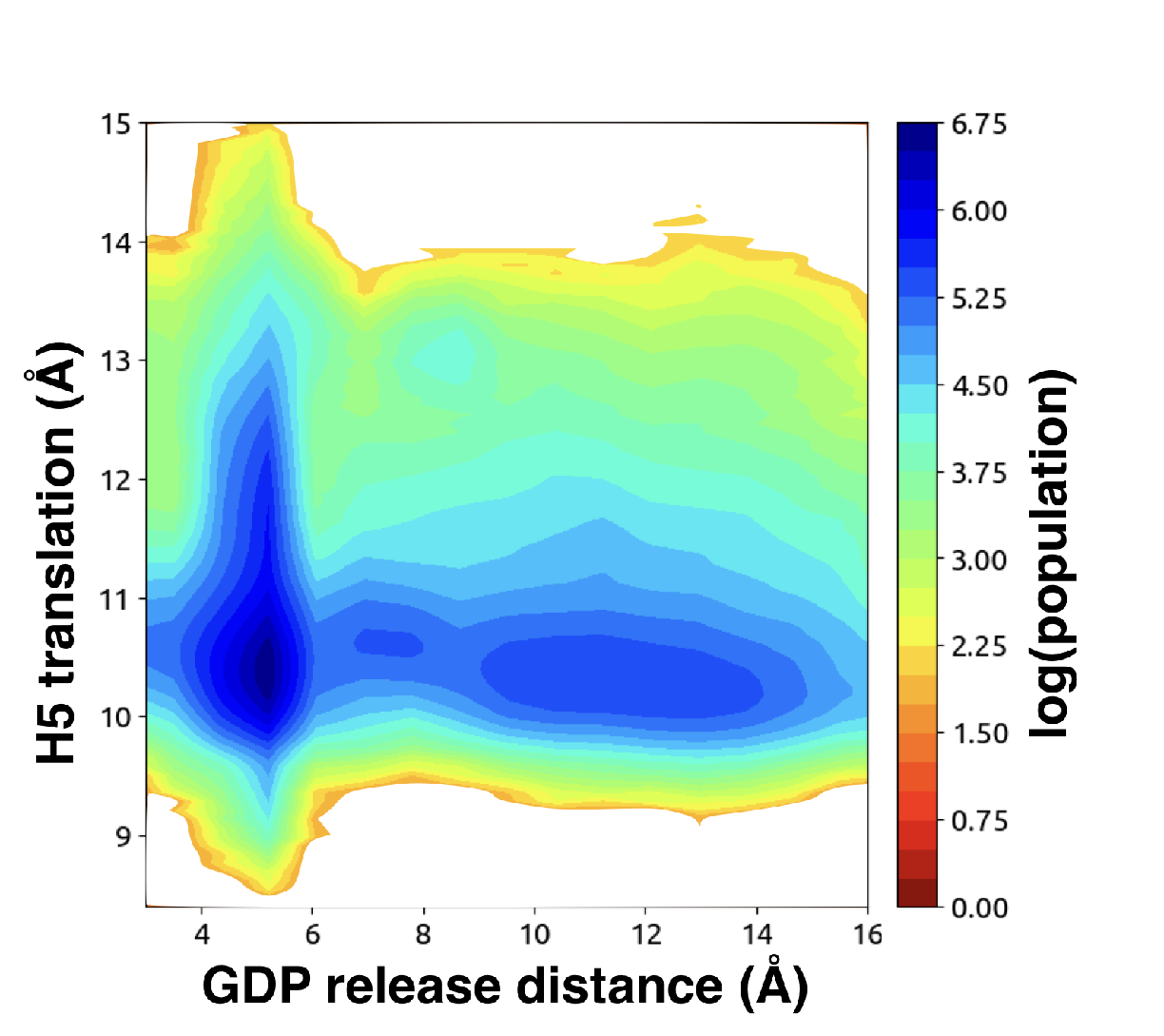
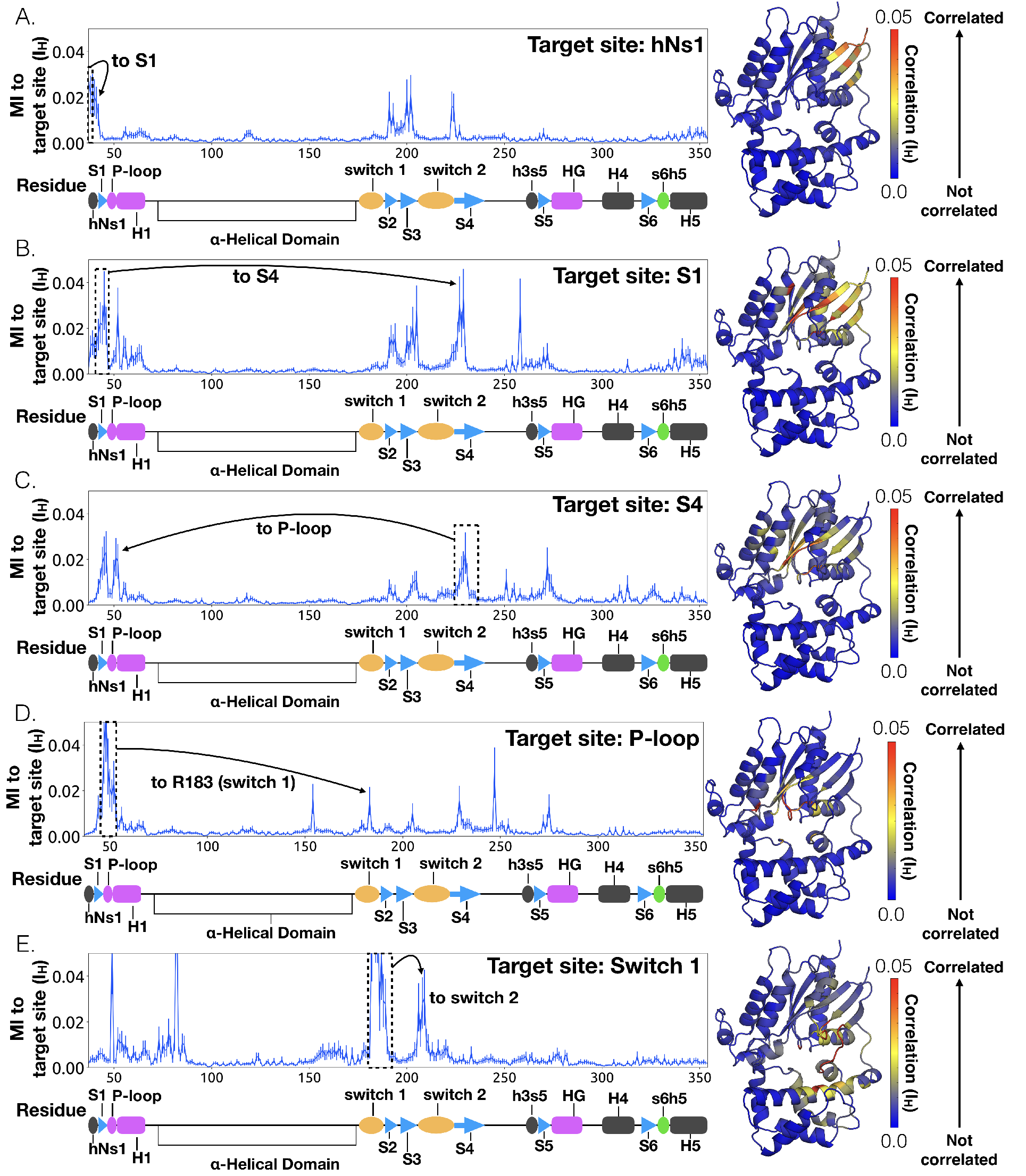
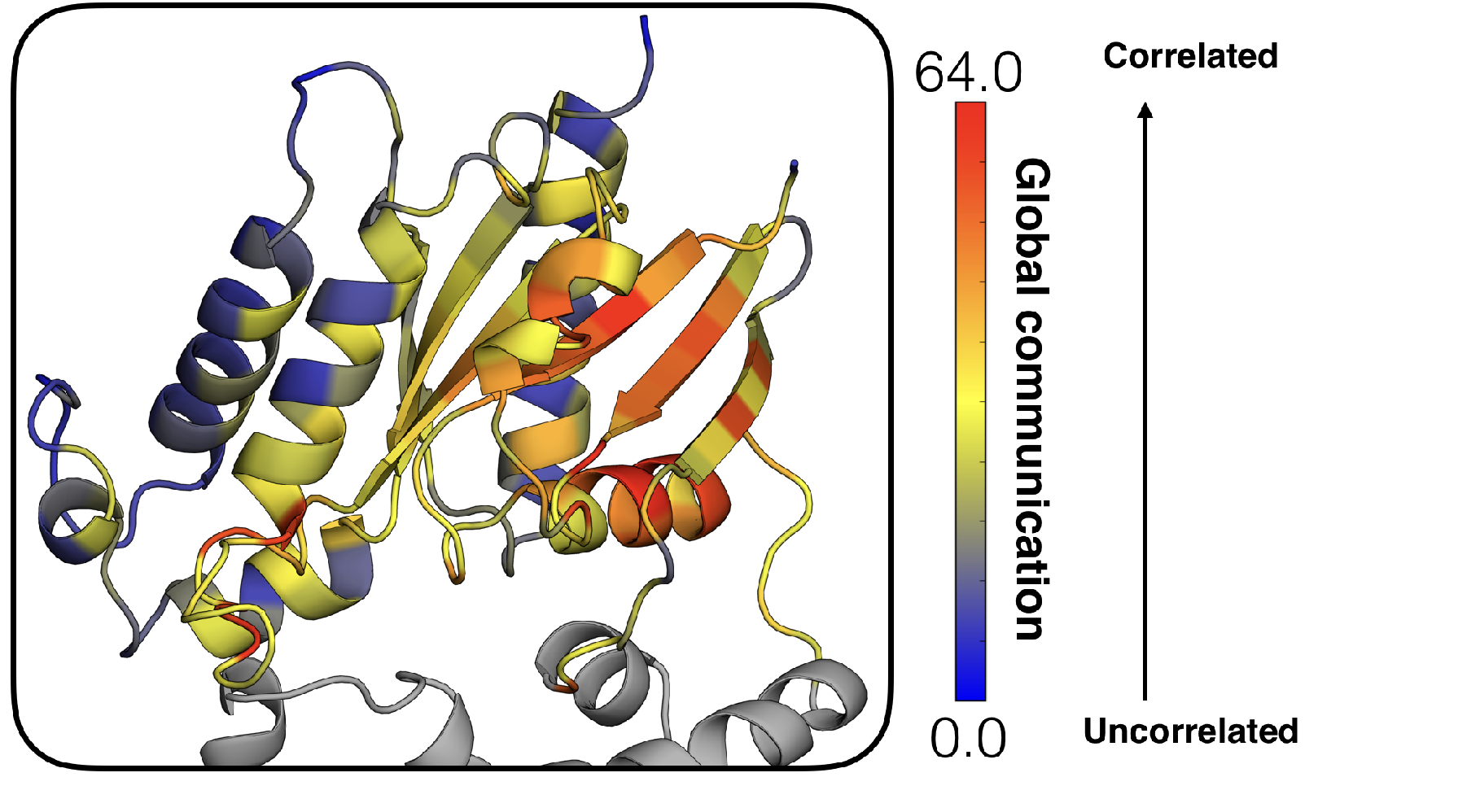
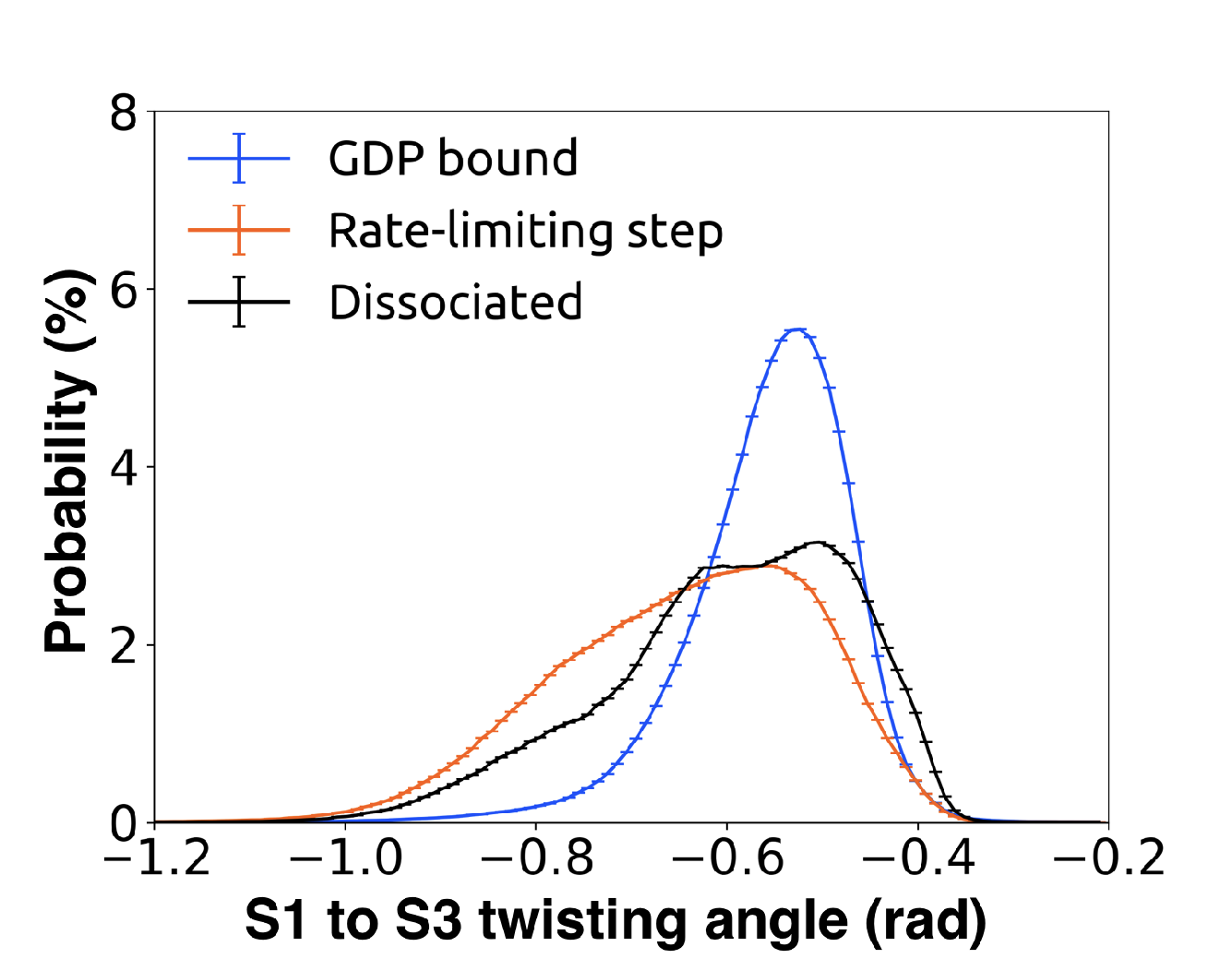
Supplementary figures highlighting the mechanism of GDP release
The work in this Appendix is published in: SSun,
X.and
Singh, S.,
Blumer, K.J., and Bowman, G.R., Simulation of spontaneous G protein activation reveals a new
intermediate driving GDP unbinding. eLife, 7, October 2018, https://doi.org/10.7554/eLife.38465.001
[48]
B.1 Supplementary Figures
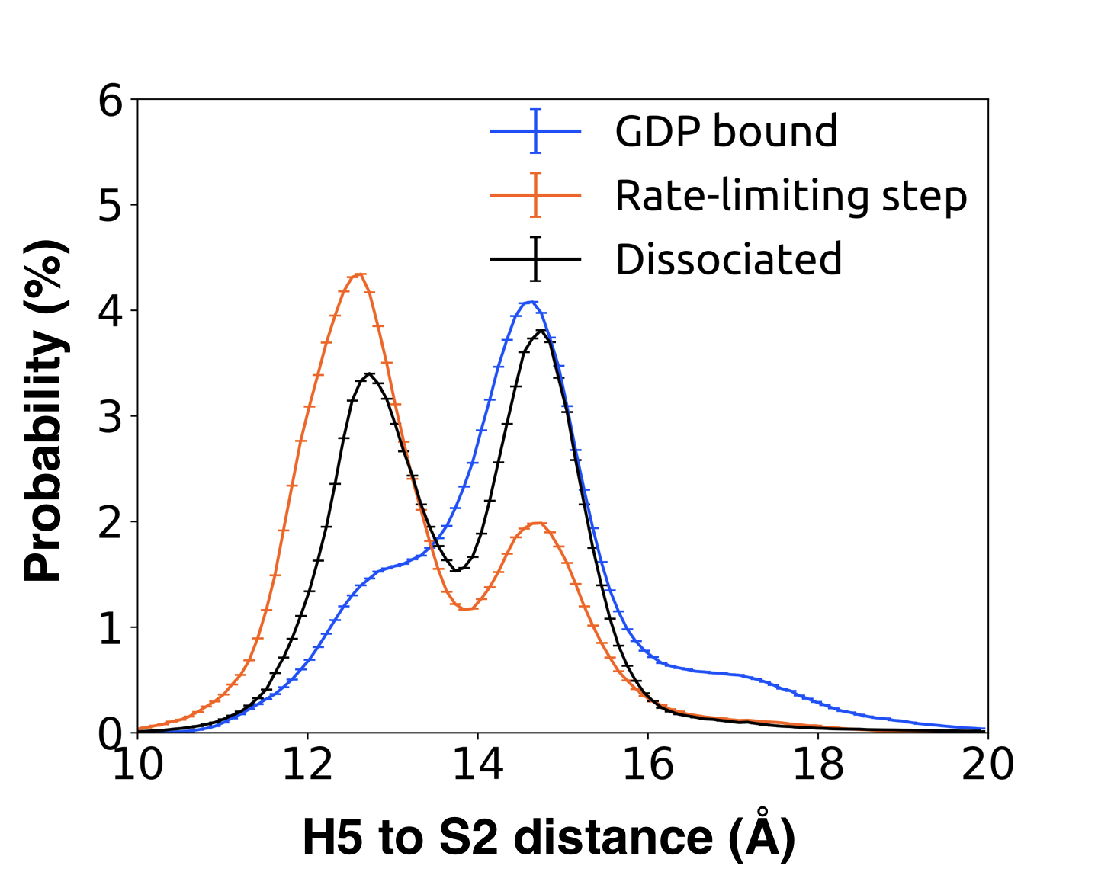
Table B.1: Measurements comparing tilting and translation of H5 across PDB structures and
MD simulation.
|
|
|
|
|
|
| Construct description | PDB ID | | | H5 Vertical | | translation distance (Å) |
| | | H5 Translation | | residues used |
|
|
|
|
|
|
|
| Gq-GDP | 3AH8 | 13.5 | 10.6 | Tyr325 to Leu349 | Thr334 to Phe341 |
|
|
|
|
|
|
| Gq after rate | | limiting step from MD |
| N/A | 15.1 | 11.1 | Tyr325 to Leu349 | Thr334 to Phe341 |
|
|
|
|
|
|
| Gi-GDP | 1GP2 | 10.3 | 10.2 | Tyr320 to Ile343 | Thr329 to Phe336 |
|
|
|
|
|
|
| Gi-µOR | 6DDF | 14.6 | 13.0 | Tyr320 to Ile343 | Thr329 to Phe336 |
|
|
|
|
|
|
| Gi-A1AR | 6D9H | 13.8 | 10.1 | Tyr321 to Ile344 | Thr330 to Phe327 |
|
|
|
|
|
|
| Gi-Rhodopsin | 6CMO | 15.8 | 10.7 | Tyr320 to Ile343 | Thr329 to Phe336 |
|
|
|
|
|
|
| Go-5HT1B | 6G79 | 13.1 | 14.2 | Tyr310 to Ile333 | Thr319 to Phe326 |
|
|
|
|
|
|
| Gs-B2AR | 3SN6 | 12.8 | 14.6 | Tyr360 to Ile383 | Thr369 to Phe376 |
|
|
|
|
|
|
| |